CBG Berry
RSP 11446
Grower: Blue Forest Farms
General Information
- Sample Name
- CBG #4
- Accession Date
- March 31, 2020
- Reported Plant Sex
- Female
- Report Type
- StrainSEEK v2 3.2Mb
- DNA Extracted From
- Stem
The strain rarity visualization shows how distant the strain is from the other cultivars in the Kannapedia database. The y-axis represents genetic distance, getting farther as you go up. The width of the visualization at any position along the y-axis shows how many strains there are in the database at that genetic distance. So, a common strain will have a more bottom-heavy shape, while uncommon and rare cultivars will have a visualization that is generally shifted towards the top.
Chemical Information
Cannabinoid and terpenoid information provided by the grower.
Cannabinoids
No information provided.
Terpenoids
No information provided.
Genetic Information
- Plant Type
- Type I
File Downloads
The bell curve in the heterozygosity visualization shows the distribution of heterozygosity levels for cannabis cultivars in the Kannapedia database. The green line shows where this particular strain fits within the distribution. Heterozygosity is associated with heterosis (aka hybrid vigor) but also leads to the production of more variable offspring. When plants have two genetically different parents, heterozygosity levels will be higher than if it has been inbred or backcrossed repeatedly.
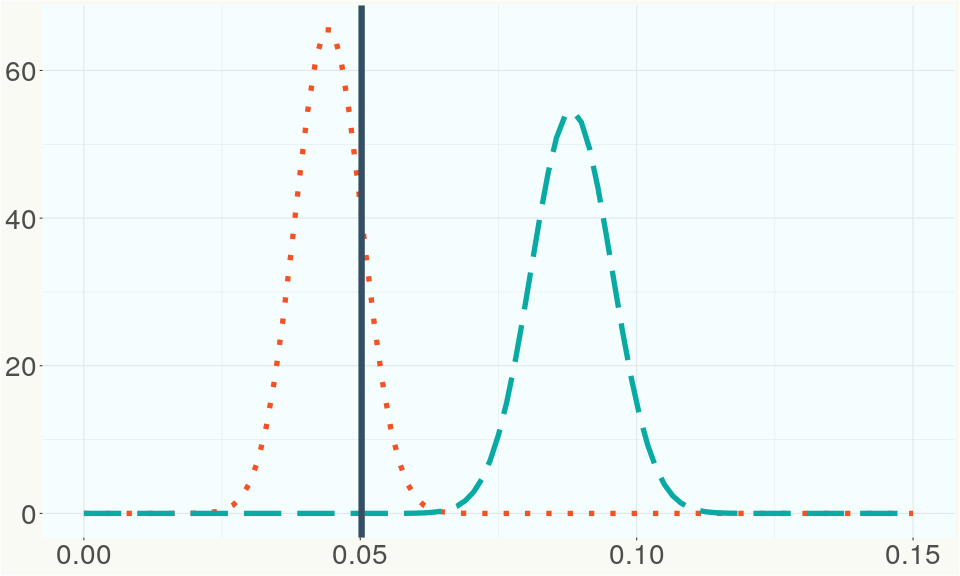
The ratio of reads mapped to Y-contigs to reads mapped to the whole Cannabis genome (Y-ratios) has been demonstrated to be strongly correlated with plant sex typing. This plot shows the distribution of Y-ratios for all samples in our database which were sequenced with the same method (panel or WGS) as this sample and where this sample falls in the distribution.
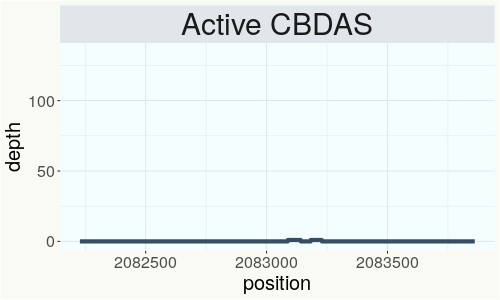
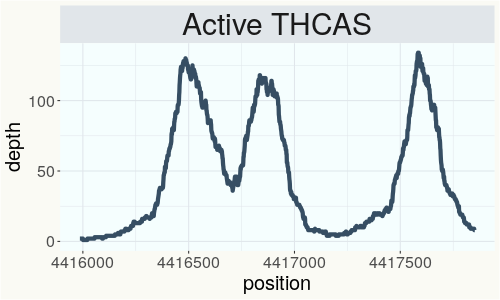
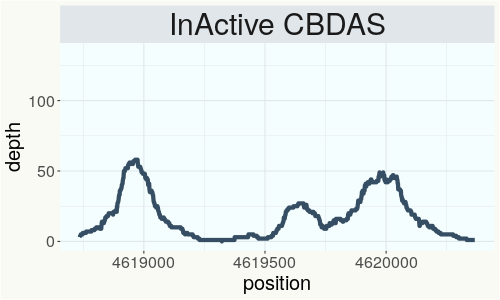
This chart represents the Illumina sequence coverage over the Bt/Bd allele. These are the three regions in the cannabis genome that impact THCA, CBDA, CBGA production. Coverage over the Active CBDAS gene is highly correlated with Type II and Type III plants as described by Etienne de Meijer. Coverage over the THCA gene is highly correlated with Type I and Type II plants but is anti-correlated with Type III plants. Type I plants require coverage over the inactive CBDA loci and no coverage over the Active CBDA gene. Lack of coverage over the Active CBDA and Active THCA allele are presumed to be Type IV plants (CBGA dominant). While deletions of entire THCAS and CBDAS genes are the most common Bt:Bd alleles observed, it is possible to have plants with these genes where functional expression of the enzyme is disrupted by deactivating point mutations (Kojoma et al. 2006).
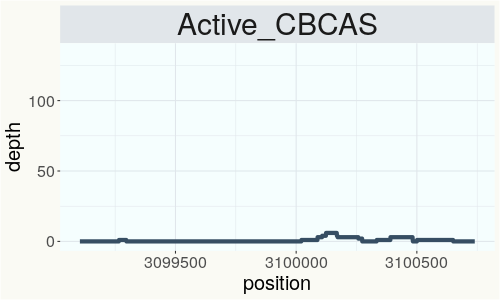
This chart represents the Illumina sequence coverage over the CBCA synthase gene.
Variants (THCAS, CBDAS, and CBCAS)
Variants (Select Genes of Interest)
| GPPs1 |
c.845_848del |
p.Glu282fs | frameshift variant | high | contig676 | 169629 | TGAAA/T |
|
| PKSG-2a | c.67T>A | p.Phe23Ile | missense variant | moderate | contig700 | 1945567 | A/T | |
| PKSG-2a | c.31A>T | p.Thr11Ser | missense variant | moderate | contig700 | 1945603 | T/A | |
| PKSG-2b | c.1152T>A | p.Asn384Lys | missense variant | moderate | contig700 | 1950486 | A/T | |
| PKSG-2b | c.1132C>G | p.Leu378Val | missense variant | moderate | contig700 | 1950506 | G/C |
|
| PKSG-2b | c.1117A>G | p.Ile373Val | missense variant | moderate | contig700 | 1950521 | T/C | |
| PKSG-2b | c.948T>G | p.Asp316Glu | missense variant | moderate | contig700 | 1950690 | A/C |
|
| PKSG-2b | c.945T>G | p.Ser315Arg | missense variant | moderate | contig700 | 1950693 | A/C |
|
| PKSG-2b | c.944G>A | p.Ser315Asn | missense variant | moderate | contig700 | 1950694 | C/T |
|
| PKSG-2b | c.934C>G | p.His312Asp | missense variant | moderate | contig700 | 1950704 | G/C |
|
| PKSG-2b | c.31A>T | p.Thr11Ser | missense variant | moderate | contig700 | 1951851 | T/A | |
| PKSG-2b | c.-2_1dupATA | start lost & conservative inframe insertion | high | contig700 | 1951880 | A/ATAT |
|
|
| DXR-2 | c.1319T>C | p.Ile440Thr | missense variant | moderate | contig380 | 285250 | A/G |
|
| aPT4 | c.97T>C | p.Tyr33His | missense variant | moderate | contig121 | 2828753 | T/C |
|
| aPT4 | c.153A>C | p.Lys51Asn | missense variant | moderate | contig121 | 2828809 | A/C |
|
| aPT4 |
c.235_236del |
p.Val79fs | frameshift variant | high | contig121 | 2829030 | ATG/A |
|
| aPT4 | c.238delT | p.Ser80fs | frameshift variant | high | contig121 | 2829034 | AT/A |
|
| aPT4 | c.302A>G | p.Asn101Ser | missense variant | moderate | contig121 | 2829099 | A/G |
|
| aPT4 | c.1168T>C | p.Tyr390His | missense variant | moderate | contig121 | 2833503 | T/C |
|
| aPT1 | c.629C>T | p.Thr210Ile | missense variant | moderate | contig121 | 2840237 | C/T | |
| HDS-2 |
c.82_93delGT |
p.Val28_Thr3 |
conservative inframe deletion | moderate | contig95 | 1989748 |
CGTAACCGGAAC |
|
| HDS-2 | c.127T>G | p.Ser43Ala | missense variant | moderate | contig95 | 1989794 | T/G |
|
Nearest genetic relatives (All Samples)
- 0.110 Doug s Varin (RSP11243)
- 0.141 CBG-#40 (RSP11444)
- 0.145 Electra (RSP11366)
- 0.154 CBG #30 (RSP11447)
- 0.156 Lift (RSP11378)
- 0.165 CBG Berry (RSP11445)
- 0.171 Joy (RSP11380)
- 0.174 UnObtanium (RSP11611)
- 0.186 Durban Poison #1 (RSP11013)
- 0.194 Suver Haze (RSP11364)
- 0.196 Serious Happiness (RSP10763)
- 0.200 Trump x Trump (RSP11466)
- 0.203 Blueberry Cheesecake (RSP10684)
- 0.206 Durban Poison #1 (RSP10996)
- 0.208 Lifter (RSP11365)
- 0.210 Liberty Haze (RSP11000)
- 0.211 Rest (RSP11377)
- 0.214 JL X NSPM1 14 (RSP11473)
- 0.214 Domnesia (RSP11184)
- 0.214 CBG Fiber (RSP11440)
Most genetically distant strains (All Samples)
- 0.446 80E (RSP11213)
- 0.437 Cherry Blossom (RSP11323)
- 0.424 80E (RSP11211)
- 0.423 80E (RSP11212)
- 0.420 R1in136 (SRR14708226)
- 0.414 Feral (RSP11206)
- 0.407 Cherry Blossom (RSP11306)
- 0.404 CS (RSP11208)
- 0.404 Feral (RSP11205)
- 0.402 Chem 91 (RSP11185)
- 0.401 VIR 223 - Bernburgskaya Odnodomnaya - bm (SRR14708217)
- 0.398 VIR 483 (SRR14708239)
- 0.397 R1in136 (SRR14708237)
- 0.397 GMO x Garlic Breath (RSP12507)
- 0.396 Delta-llosa (SRR14708272)
- 0.395 Juso14 (SRR14708259)
- 0.395 Feral (RSP10890)
- 0.394 Chematonic -Cannatonic x Chemdawg- (RSP11394)
- 0.394 Feral (RSP10892)
- 0.394 Carmagnola (RSP11037)
Nearest genetic relative in Phylos dataset
- Overlapping SNPs:
- 81
- Concordance:
- 48
Nearest genetic relative in Lynch dataset
- Overlapping SNPs:
- 6
- Concordance:
- 6
Blockchain Registration Information
- Transaction ID
-
9aff1861bdd1afc9
8890204fce9563e4 5a434667b1870b6c 9323cb61db18d4e8 - Stamping Certificate
- Download PDF (39.9 KB)
- SHASUM Hash
-
0f99fa84bec6bb974e892b34c21906f9 cf20483bfd815d86 e945c562b97d2fd2